Building a Generalizable Model for Ni-Photoredox Cross-Electrophile Coupling
Active‑learning yield model maps 22k+ Ni/photoredox cross‑electrophile couplings with <400 HTE data points, fusing DFT features and uncertainty querying to pred
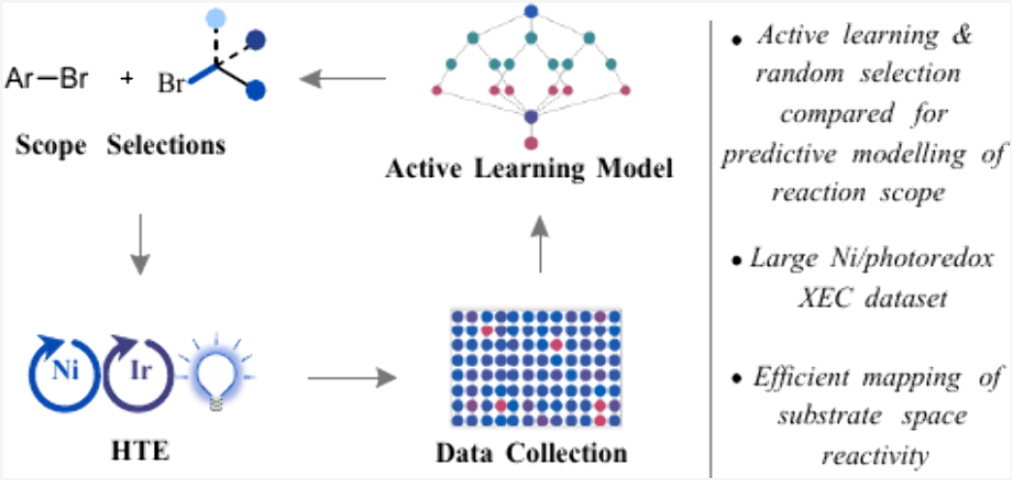
When developing machine learning models for yield prediction, the two main challenges are effectively exploring condition space and substrate space. In this article, we disclose an approach for mapping the substrate space for Ni/photoredox-catalyzed cross-electrophile coupling of alkyl bromides and aryl bromides in a high-throughput experimentation (HTE) context. This model employs active learning (in particular, uncertainty querying) as a strategy to rapidly construct a yield model. Given the vastness of substrate space, we focused on an approach that builds an initial model and then uses a minimal data set to expand into new chemical spaces. In particular, we built a model for a virtual space of 22,240 compounds using less than 400 data points. We demonstrated that the model can be expanded to 33,312 compounds by adding information around 24 building blocks (<100 additional reactions). Comparing the active learning-based model to one constructed on randomly selected data showed that the active learning model was significantly better at predicting which reactions will be successful. A combination of density function theory (DFT) and difference Morgan fingerprints was employed to construct the random forest model. Feature importance analysis indicates that key DFT features that are related to the reaction mechanism (e.g., alkyl radical LUMO energy) were crucial for model performance and predictions on aryl bromides outside the training set. We anticipate that combining DFT featurization and uncertainty-based querying will help the synthetic organic community build predictive models in a data-efficient manner for other chemical reactions that feature large and diverse scopes.





